The Laboratory of Electron Transport Membrane Proteins and Structural Bioenergetics
We are interested in understanding the mechanism of electron transport membrane proteins, a varied class of enzymes essential for energy transduction and cellular defense/signaling. We research both the basic biology of these enzymes and translational applications to human health and disease.
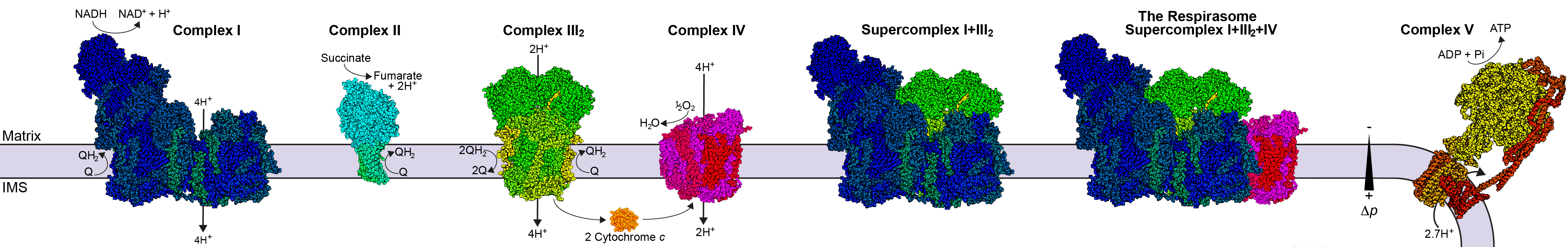
All cells, including human pathogens, store energy in the form of a proton gradient across a membrane. The transport of electrons from high-energy donors (e.g., NADH or succinate) to lower energy acceptors (e.g., molecular oxygen, O2) is coupled to proton pumping by essential membrane protein complexes. The electron transport chain complexes of aerobic organisms generally fall into five groups: proton-pumping NADH-coenzyme Q oxidoreductases (complex I), non-proton-pumping coenzyme Q reductases (complex II), proton-pumping coenzyme Q-cytochrome c oxidoreductases (complex III), proton-pumping oxidases (complex IV) and ATP synthase (complex V). In some organisms, these complexes associate to form higher-order supercomplexes whose physiological roles remain enigmatic, but are thought to include substrate channeling and the regulation of reactive oxygen species (ROS) production.
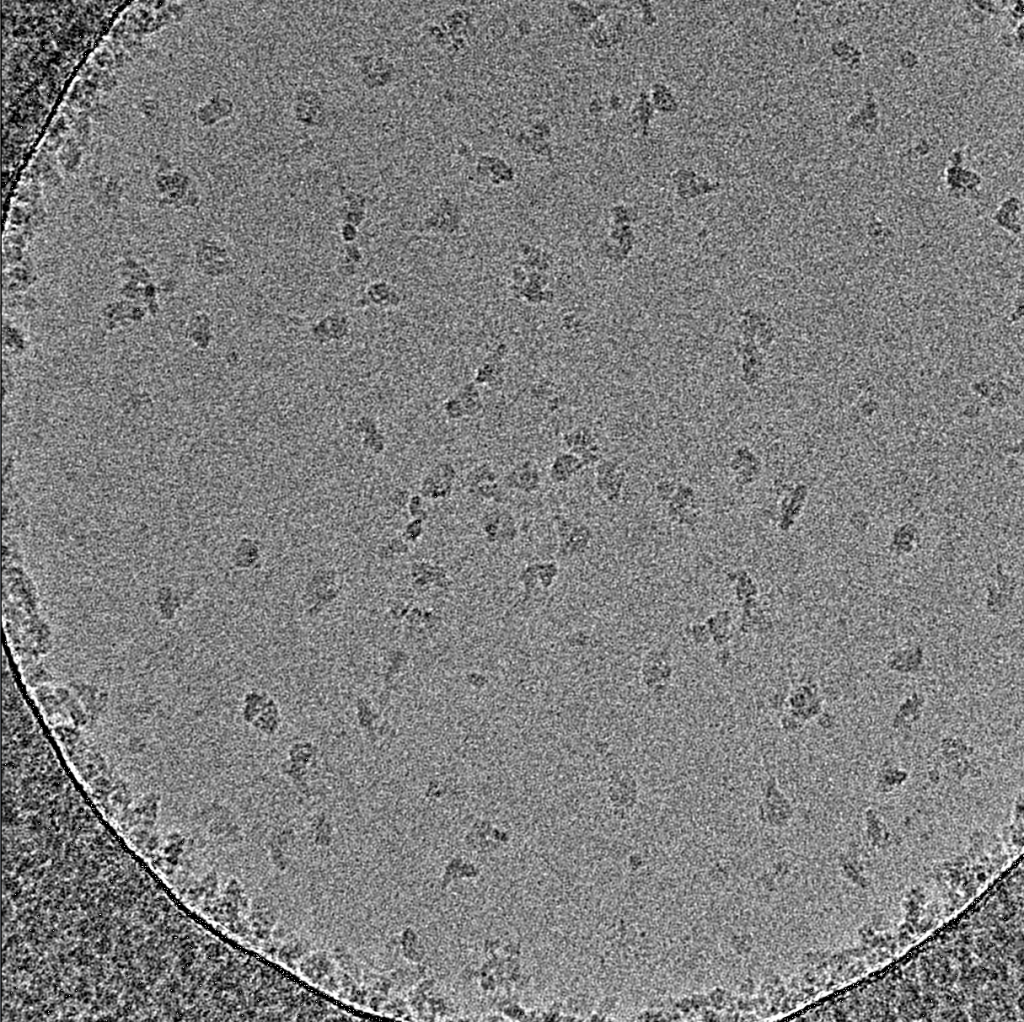
Our lab uses single-particle electron cryo-microscopy (cryo-EM) together with other biochemical, biophysical and structural methods to elucidate the mechanism of electron transport proteins involved in energy transduction and cellular defence/signalling.
Current Research:
Diversity of Eukaryotic Core Respiratory Metabolism: A chain of multi-subunit protein complexes in the inner mitochondrial membrane of eukaryotes catalyze the core biological energy-converting process known as cellular respiration or oxidative phosphorylation. Much of what we know about this key biological process at the molecular scale comes from a small set of model organisms in the Opisthokont (fungi and metazoans) branch of the eukaryotic tree of life. Our working hypothesis in the lab is that these standard model organisms under-represent the biological diversity of this critical metabolic pathway and that by studying the existing diversity across eukaryotes will reveal core aspects of conserved mechanism as well as new ways to modulate the activity of the complexes.
Given their distinct mitochondrial morphology and divergent ATP synthase complex, we reasoned that the respiratory complexes of the model ciliate Tetrahymena thermophila (part of the SAR super group of eukaryotes) would be different from those of the model Opisthokonts. Our structures of the T. thermophila respiratory complexes revealed this diversity and forced us to question much of what we thought was invariant in respiration as well as providing new insight into the physiological function of respiratory supercomplexes.
This movie shows the cryogenic electron microscopy reconstruction of the T. thermophila respiratory SC I+III2 colored by subunit.
This movie shows the cryogenic electron microscopy reconstruction of the T. thermophila respiratory CIV2 colored by subunit.
Plant Respiration: Despite the centrality of respiration to plant biomass accumulation, carbon flux and acclimation, the fundamental mechanisms plant mitochondrial complexes use to produce electrochemical proton gradients that are converted into chemical energy remain unknown. A detailed functional and structural understanding of plant respiratory mitochondrial electron transport chain (mETC) complexes is essential to understand the fundamental mechanisms of bio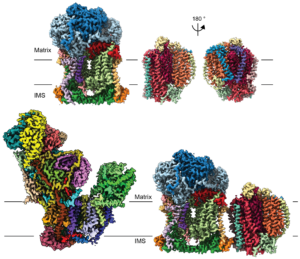 logical energy conversion. By applying expertise in mitochondrial-membrane-protein biochemistry and structure biology, we recently obtained cryoEM structures of the protein complexes CI*, CIII2, CIV and SC III2+IV from Vigna radiata (mung bean)—the first high-resolution structures for mitochondrial complexes and supercomplexes for any plant. We are continuing to characterize the plant-specific features of these and other plant mitochondrial respiratory complexes and supercomplexes functionally and structurally, using biochemical and biophysical approaches including chromatographic purifications, single particle cryoEM and enzyme-kinetic analyses.
logical energy conversion. By applying expertise in mitochondrial-membrane-protein biochemistry and structure biology, we recently obtained cryoEM structures of the protein complexes CI*, CIII2, CIV and SC III2+IV from Vigna radiata (mung bean)—the first high-resolution structures for mitochondrial complexes and supercomplexes for any plant. We are continuing to characterize the plant-specific features of these and other plant mitochondrial respiratory complexes and supercomplexes functionally and structurally, using biochemical and biophysical approaches including chromatographic purifications, single particle cryoEM and enzyme-kinetic analyses.
A deep structural/functional understanding of the components of plant mETC will provide insights into bioenergy conversion. Our research will test hypotheses generated by the recent structures and provide answers to longstanding mechanistic questions such as the catalytic function of CI’s carbonic anhydrase domain and the interplay between CIII2’s dual roles (respiration and mitochondrial peptide processing). We are also test novel hypotheses triggered by the structures, such as the potential functional role of CI*. These studies will likely produce additional breakthrough structures that will enable new detailed hypotheses regarding the mechanisms of these plant complexes and supercomplexes. The project will shed light on the fundamental tenets of electron transport in plant mitochondria and will generate approaches, materials and hypotheses for the continued mechanistic examination of energy-converting enzymes in plants, advancing the field of plant respiration and bioenergetics. The research can also further our understanding of other areas of plant biology such as photosynthesis, biomass accumulation and stress responses.
This work is supported by a DOE Early Career Research Award from the Office of Basic Energy Sciences.
Read more about it with eLife Digests:
The complex story of plant resipiration
Past Research: Membrane Protein Structure/Function
Throughout his training, Dr. Letts has combined structural approaches with functional and biophysical techniques to understand the molecular mechanisms underlying the biological function of membrane and soluble proteins. As an undergraduate, Dr. Letts got involved in research early on by joining the X-ray crystallography laboratory of Dr. Stephen Evans at the University of Victoria, where he studied the structures of the human ABO(H) blood-group glycosyltransferase enzymes. There he initially learned the methods of molecular biology, protein purification, crystallization and X-ray crystallographic structure determination. For his doctoral research, Dr Letts joined the laboratory of Nobel Laureate Dr. Roderick MacKinnon at Rockefeller University. There, he continued using X-ray crystallography and also learned several new techniques related to membrane protein research: expression in bacteria, yeast and mammalian cells, purification, reconstitution and functional characterization, as well as electrophysiology and NMR. As a postdoctoral fellow in Dr. Leonid Sazanov’s group, Dr. Letts gained extensive experience in the purification of large membrane protein complexes from natural source. For structural characterization, he learned and became proficient in single-particle cryo-EM.
As an independent investigator, Dr. Letts plans to combine single-particle cryo-EM and membrane protein reconstitution to study the structure and function of electron transport membrane proteins.
Postdoctoral research: Dr. Letts’ postdoctoral studies in the laboratory of Dr. Leonid Sazanov focused on the determination of the atomic structures of large membrane-protein complexes and supercomplexes of the mitochondrial electron transport chain.
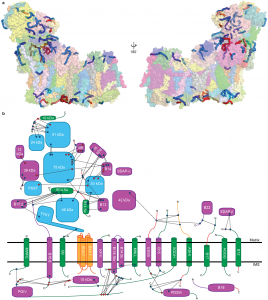
Specifically, he developed a novel protocol for the purification of complex I from sheep-heart mitochondria that results in a highly stable and active complex. Using this protocol, in collaboration with a graduate student in the lab Karol Fiedorczuk (now Dr. Fiedorczuk), he were able to solve the first atomic structure of complex I by single-particle cryo-EM. In conjunction with cross-linking mass spectrometry, this structure revealed the position of each of the 31 supernumerary subunits and the details of their interactions within the full complex at 3.9 Å resolution. This structure provides the framework for the mechanistic understanding of the coupling between electron transfer and proton pumping in complex I and for the modelling of known disease-causing mutations. Additionally, the identification of several bound co-factors and lipids shed light on and generated new hypotheses for the diverse regulatory mechanisms of the complex.
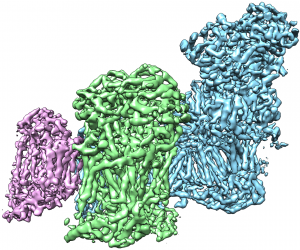
In parallel, Dr. Letts solved the architecture of the respiratory supercomplex formed by complex I, complex III and complex IV (the “respirasome”) in two states, as well as the architecture the supercomplex formed between complex I and complex III alone. These architectures at 5.4-7.8 Å revealed details of inter-complex interactions within the supercomplexes, demonstrated the lack of constrained substrate channels, provided insight into the possible mechanism of supercomplex stabilization by putative assembly factors and led to the proposal of a mechanism for the reduction of reactive oxygen species (ROS) generation. This investigation on respiratory supercomplex structure and function continues via the characterization of a functional supercomplex I-III2 using a novel purification protocol, which has resulted in maps at better than 4 Å resolution.
Ph.D. research: Dr. Letts’ doctoral research focused on structural and functional studies of the human voltage-gated proton channel (hHV1). By expressing the channel in HEK cells and performing site-specific cross-linking in the membrane, he, together with post-doc collaborator Dr. Seok-Yong Lee, demonstrated that this channel is a functional dimer. Furthermore, through expression of the channel in Pichia pastoris, purification and reconstitution, they demonstrated that the putative channel gene is able to conduct protons independently of any cellular factors. Given that raising antibodies against the membrane portion of the channel for anti-body mediated crystallization proved intractable, a chimeric approach was taken in which Dr. Letts spliced a known antibody epitope of a homologue into the voltage-sensor domain. In this way, he was able to obtain diffraction-quality crystals of an hHV1-Fab complex and solve their structure. However, the maps revealed that the channel in the crystal was in a non-physiological, partially unfolded state. Thus, Dr. Letts decided to adopt a different approach and, in collaboration with Dr. Joel Butterwick, attempted to solve the structure of the hHV1 voltage-sensor domain using NMR. Although they were able to clearly define the secondary structure of the protein, they again found that the protein became partially unstructured in detergent. Given that each construct was functional upon reconstitution, these studies revealed the importance of the membrane bilayer for the stabilization of this class of membrane proteins and the importance of structure validation using biochemical and biophysical approaches.